New Mathematics Advances the Frontier of Macromolecular Imaging
Berkeley Lab’s M-TIP solves the reconstruction problem for fluctuation X-ray scattering
August 10, 2015
Contact: Linda Vu, +1 510 495 2402, lvu@lbl.gov
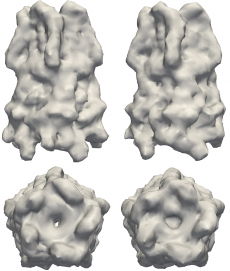
Structures to the left are models of the pentameric ligand-gated ion channel (pLGIC), which mediate fast synaptic communication by converting chemical signals into an electrical response. The structures on the right are reconstructions of pLGIC from FXS data using M-TIP. (Image Credit: Jeffrey J. Donatelli, Berkeley Lab)
A comprehensive understanding of complex nanostructures—like proteins and viruses—could lead to breakthroughs in some of the most challenging problems in biology and medicine. But because these objects are a thousand times smaller than the width of human hair, scientists can’t directly see into them to determine their shape and function.
In order to visualize the structure of proteins in their native environment, scientists can blast powerful X-ray beams at tiny volumes of proteins in solution. The resulting ‘diffraction patterns’ can then be interpreted to determine, or reconstruct, information about the protein’s molecular structure. However, traditional solution scattering techniques are often limited by how much structural detail they can extract. As brighter and more powerful light sources come online, researchers are devising new ways to overcome these limitations. In particular, an emerging technique known as fluctuation X-ray scattering (FXS) could provide orders of magnitude more detail than traditional solution scattering. Until now, a major limitation for FXS has been a lack of mathematical methods to efficiently interpret the data.
In a paper published August 3, 2015 in the Proceedings of the National Academy of Sciences, Lawrence Berkeley National Laboratory (Berkeley Lab) applied mathematicians Jeffrey Donatelli and James Sethian, and physical bioscientist Peter Zwart have introduced new mathematical theory and an algorithm, which they call "Multi-tiered iterative phasing (M-TIP)," to solve the reconstruction problem from FXS data. Their code can quickly determine general structure in only a few minutes on a desktop computer. This approach is an important step in unlocking the door to new advances in biophysics and has the promise of ushering in new tools to help solve some of the most challenging problems in the life sciences.
"These are exciting times,” says Zwart, who is a member of the Physical Biosciences Division at Berkeley Lab. "Although fluctuation scattering was first proposed 38 years ago, its routine practical realization has only now become feasible with the advent of modern X-ray light sources. This novel reconstruction method plays a central role in mapping out the strengths of fluctuation scattering as a routine biophysical technique."
Overcoming the Limitations of Traditional Imaging
With advances in light source technology, diffraction can be done in a many different ways. If particles can be organized into sufficiently large crystals, their structure can be determined through crystallography, which involves shooting x-rays through a crystal. But, many important structures are too floppy to succumb to crystallization and may have a different structure in solution compared to what is determined from crystallography.
As an alternative and complementary technique, structural biologists often gather diffraction patterns from particles in solution. However, in these so called small- and wide-angle x-ray scattering (SAXS/WAXS) experiments, particles can rotate during imaging, which results in a loss of information and often leads to a poor reconstruction of the unknown structure. This is analogous to a photo of a child on a merry-go-round taken with an exposure time that is too long: the picture is blurred and details are lost.
One solution to overcome the limitations of SAXS/WAXS is to build faster and brighter light sources that can "take a picture" far quicker than this rotational diffusion time. This is the promise of new DOE facilities relying on free electron lasers (FEL), such as the Linac Coherent Light Source (LCLS) at Stanford. Extracting additional angular correlation information through a technique known as fluctuation X-ray scattering (FXS) has been suggested in order to reconstruct more structural details of the imaged object. However, solving the inverse problem of using this type of information to determine molecular structure is challenging, and until now has been a major stumbling block.
Deciphering FXS with M-TIP

Illustration of a fluctuation scattering experiment, which collects X-ray diffraction snapshots of multiple identical proteins in solution. An ultrabright X-ray laser provides fast snapshots which avoids motion blur, ultimately resulting in a detailed view of biological nanostructures. (Image Credit: Peter Zwart, Berkeley Lab)
Part of the challenge of generating a model from fluctuation scattering data stems from the fact that, unlike in standard diffractive imaging which measures diffraction intensities and only requires recovering the missing complex phases, inverting FXS data requires the recovery of the three-dimensional intensity information as well.
The team’s new "M-TIP" algorithm alternatingly projects a model to agree with the FXS data along with any prior known constraints about the solution, such as density upper and lower bounds, size, and/or symmetry, and can simultaneously determine the intensities, complex phases, and molecular structure.
“In order to develop a robust and efficient FXS reconstruction algorithm, we had to solve a number of non-trivial mathematical problems,“ says Donatelli of Berkeley Lab’s Computing Sciences Division. “Deriving the relation between structure and FXS data involves a substantial amount of harmonic analysis and linear algebra and we also needed to develop several new computational tools, such as a fast and reliable polar Fourier transform.”
Given that FXS is still a relatively new technique, there are no experimental data sets in the public domain. To compensate for the lack of such data, Donatelli, Sethian and Zwart tested their method on simulated FXS data on various test shapes, including a model of a pentameric ligand-gated ion channel (pLGIC). Their "M-TIP" algorithm was able to quickly produced accurate, high-resolution reconstructions of these shapes from their corresponding FXS data.
CAMERA: Innovation through Cross-Disciplinary Science
This work is part of a new project being undertaken by CAMERA (The Center for Advanced Mathematics for Energy Research Applications). CAMERA is a joint effort between DOE’s Office of Advanced Scientific Computing Research and Office of Basic Energy Sciences. Led by Sethian, CAMERA brings together mathematicians, experimental scientists, computer scientists, and software engineers to develop and deliver new mathematical tools and software to data and imaging challenges at the DOE facilities, including work at synchrotron light sources and nanoscience research centers.
"DOE’s light sources offer a rich environment for tackling wonderful math problems whose solutions can make a major impact on fast moving science'' says Sethian. "Combining Zwart's insight into the problem with Donatelli's background in harmonic analysis and iterative phasing algorithms set the stage for a new way to think about reconstruction from FXS data."
The Future of FXS
Beamtime at the LCLS was recently awarded to the authors as part of a large multi-institutional collaboration to collect FXS data from several different biological specimens. This will allow the researchers the opportunity to test and, if necessary, tune their reconstruction techniques on experimental data.
"Ultimately, the goal is to provide the scientific community with a powerful new tool to determine the structure and dynamics of nano-sized particles in a routine, high throughput fashion," says Zwart. "The full deployment of FXS as a new tool in the arsenal of the structural biologist will take some time, but this is an important breakthrough."
The researchers emphasize that FXS data may also be collected using an ultrabright synchrotron light source from particles cryogenically frozen in place. The National Institutes of Health recently awarded Zwart and coworkers a new detector for the development of this method on synchrotron light sources.
"Recent advances in detectors, X-ray sources and optics bring fluctuation scattering of cryogenically frozen large macromolecular machines within practical reach of modern synchrotrons," says Steven Kevan, Deputy Director for Science at the Berkeley Lab’s Advanced Light Source (ALS). "We are looking forward to the development of this technique at the Advanced Light Source."
The researchers note that although the method they developed has been applied on problems specific to the life sciences, it can be extended to other applications in material and energy sciences as well. The work was supported by DOE’s Office of Science (Office of Advanced Scientific Computing Research and Office of Basic Energy Sciences) and by the National Institute of Health. The Advanced Light Source is an Office of Science User Facility.
About Berkeley Lab
Founded in 1931 on the belief that the biggest scientific challenges are best addressed by teams, Lawrence Berkeley National Laboratory and its scientists have been recognized with 16 Nobel Prizes. Today, Berkeley Lab researchers develop sustainable energy and environmental solutions, create useful new materials, advance the frontiers of computing, and probe the mysteries of life, matter, and the universe. Scientists from around the world rely on the Lab’s facilities for their own discovery science. Berkeley Lab is a multiprogram national laboratory, managed by the University of California for the U.S. Department of Energy’s Office of Science.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science.









