Computational Analysis Enables Breakthrough in Biomolecular Dynamics
Scientific Data Division Researchers Use Machine Learning Methods to Develop Semi-Automated Analysis Pipeline
April 20, 2022
By Carol Pott
Contact: cscomms@lbl.gov
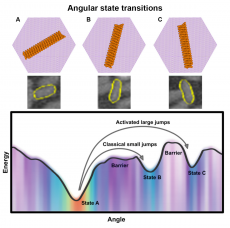
Schematic of angular states and HS-AFM snapshots of protein nanorods in their energetically preferred orientations, corresponding to specific directions of the mineral lattice. Orientational free energy landscape and heat map of relative populations at each angle determined from deep learning analysis of HS-AFM data. (Credit: Stephane A. King, PNNL)
Living systems create a wide range of biomolecular arrays with complex functions and exquisite organization. This has inspired synthetic equivalents for a range of applications, enabling new high-throughput approaches to biocomposites, diagnostics, and materials research. Although past studies have revealed complex modes of nanoparticle motion, until this breakthrough a systematic picture of the energetic controls and response dynamics leading to that organization was elusive.
A new study with data analyses from Lawrence Berkeley National Laboratory (Berkeley Lab) computational researchers helps broaden the physical understanding of biomolecular assembly by tracking motion at unprecedented resolution and defining a general procedure for using in situ visualization and machine learning to explore such dynamics. This research characterizes the “energy landscape” for protein orientation and, by analyzing the motion of the proteins, shows how that energy landscape controls the rate of motion between different orientations.
Published April 11, 2022, in the Proceedings of the National Academy of Sciences (PNAS), Berkeley Lab Scientific Data Division (SDD) researchers E. Wes Bethel, Talita Perciano, and Oliver Rübel joined a collaborative team of researchers from Pacific Northwest National Laboratory, and the University of Washington to develop deep learning techniques for analyzing in situ high-speed atomic force microscopy (HS-AFM) and transition electron microscopy data.
The project studied the orientational behavior and energy landscape of protein nanorods as they transition through energy states, ultimately ending up in some stable energetically favorable orientation. Protein nanorods are incredibly small (just a few tens of a nanometer long) and they move very quickly, making it extremely hard to study their motion over time. This required techniques that could image the proteins in water at extremely high speeds (less than one second per frame), with sub-nanometer resolution. The imaging produced an inordinate amount of data, so the Berkeley Lab team used deep learning-based image analysis and Monte Carlo simulations to investigate the rotational dynamics of these rod-like proteins on a mineral surface.
“Before the SDD team got involved with the project, the researchers were only able to observe the behavior of one or two of these nanorods over time,” said Bethel, a senior computer scientist in the SDD. “We developed an analysis pipeline using machine learning methods that solved that problem and enabled semi-automated analysis of thousands of nanorods at a time, dramatically decreasing the time needed to process the images and allowing for increased insight into the protein motion.”
Computational analyses of the data were performed by SDD researchers, who created a multi-step image analysis workflow that included denoising, deep learning segmentation, tracking, and Markov model analysis. These tools were critical to extracting the cooperative behavior of the system and identifying the physical mechanisms through comparison with the simulations.
The first step in the pipeline involved segmenting nanorods using a deep neural network architecture, U-NET, commonly used for microscopy and medical image segmentation. Deep learning segmentation often contends with the challenge of separating objects in cluttered microscopy images. The high level of noise makes it difficult to detect boundaries. To combat this, the team used an additional neural network trained at the center of the rods to get better segmentation, detect objects and label them, and precisely outline each unique shape.
Bilateral filtering and contrast limited adaptive histogram equalization (CLAHE) were used for the preprocessing step to correct unbalanced illumination, reduce noise, and improve the contrast of the input images. The bilateral filter smoothed uniform regions of the image while preserving details such as the edges and borders of the objects. After improving the image quality with the bilateral filter, the next step was to light-correct and emphasize targets for segmentation by applying the CLAHE method, which uses information based on histograms computed over different regions of the image. The result enhanced local details even in regions that were darker or lighter than the rest of the image.
“During the image-analysis pipeline development, we evaluated the impact of the noise and the light imbalance from the images on the final segmentation results,” said Perciano, an SDD research scientist. “As a result, we discovered that we could reduce noise and make the regions of the rods clearer by applying these preprocessing techniques, leading to better segmentation accuracy.”
With the strong results from the segmentation process, tracking was performed using a graph-based method to maximize the amount of data extractable from any given frame. Again, the noisy, cluttered images presented a challenge.
“This work closes the loop between simulations and experimental data,” said Rübel, also an SDD research scientist. “We are comparing kinetic Monte Carlo simulations to experimental Markov models, using the results from those experiments to better understand the theoretical model behind the rotational dynamics of these nanorods.”
Using the segmentation results, the team performed Markov model analysis of frame-to-frame data associations, allowing them to find the angles that the rods tend to prefer and creating a statistical Markov-based model displaying how the rods move within one-second intervals. By quantifying the rotational dynamics of de novo-designed proteins on the mica lattice, the team simultaneously determined the orientational energy landscape and transition probabilities between energetically favorable orientations.
“This was an important opportunity to advance computational capabilities for experimental and observational science,” said Deb Agarwal, interim division director, SDD. Starting in the winter of 2017, the Department of Energy’s Basic Energy Science and Advanced Scientific Computing Research-funded research features a unique combination of an Energy Frontier Research Center (EFRC) Center for the Science of Synthesis Across Scales team with the Scientific Discovery through Advanced Computing (SciDac) Institute for Resource and Application Productivity through Computation, Information, and Data Science (RAPIDS) team.
About Berkeley Lab
Founded in 1931 on the belief that the biggest scientific challenges are best addressed by teams, Lawrence Berkeley National Laboratory and its scientists have been recognized with 16 Nobel Prizes. Today, Berkeley Lab researchers develop sustainable energy and environmental solutions, create useful new materials, advance the frontiers of computing, and probe the mysteries of life, matter, and the universe. Scientists from around the world rely on the Lab’s facilities for their own discovery science. Berkeley Lab is a multiprogram national laboratory, managed by the University of California for the U.S. Department of Energy’s Office of Science.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science.









